A wider and more diverse world of genomes:Closing in on the mystery of evolution by making full use of state-of-the-art analysis technologies

Kanako Koyanagi, Doctor of Science,
Associate Professor of the Graduate School of Information Science and Technology, Hokkaido University,
Division of Bioengineering and Bioinformatics’s Research Group of Bioinformatics
Profile:
Associate Professor Koyanagi finished her doctoral program at Kyoto University Graduate School of Science. She became involved in the construction of an integrated human gene database at the Japan Biological Information Research Center in 2001 and became part of an international joint research project (H-Invitational) aimed at constructing an integrated database of human genes in 2002. In the project she conducted comparisons of human full-length cDNAs and genome sequences, performed analysis, and organized genetic information. She carried out annotation of approximately 40,000 full-length cDNAs in collaboration with researchers from Japan and abroad. After working as Assistant Professor at the Nara Institute of Science and Technology in 2003, she joined Hokkaido University Graduate School of Information Science and Technology as Associate Professor in 2004. Currently, she is working on the elucidation of the evolution of genetic information in humans, viruses, and other organisms through computer analysis of their genomes, among other research topics.
Dramatic expansion of research field and application scope through innovations in technology
What kinds of research are conducted in the field of genome research today?
Dr. Koyanagi: Genome refers to the total genetic information (DNA) found in a cell of a living organism. The human genome is made up of around three billion nucleotides (of four base types, namely, adenine (A), guanine (G), cytosine (C), and thymine (T)) that include approximately 22,000 genes. The sequencing of the entire human genome, which was a huge project involving research institutions around the world and required many years and several billions of dollars in costs, was completed in 2003. Since then, rapid advances in technological innovation have taken place, such that human genomes can now be sequenced in a matter of days and costs of only a few thousand dollars.
With the ease by which genomes can now be sequenced, the scope and applications of genome research have substantially expanded. The function of human genes in the human body and the particular timing are gradually becoming clear. It is also now possible to sequence a person’s individual genome and determine an individual’s predisposition to particular diseases based on genetic information.
However, at this point, we have only come to elucidate the functions and roles of around half of all human genes, meaning that we still have a lot of ground to cover. The ability to overcome technical barriers to the study of human genomes and to analyze them at significantly lower cost and shorter time has hastened the speed of research and led to many new discoveries, and at the same time brought to the fore many new questions and topics. The body of knowledge about genomes is indeed growing at an explosive pace.
Inferring evolutionary processes from genetic differences
What kinds of research programs are you pursuing at your laboratory amidst this rapidly changing environment surrounding genome research?

Dr. Koyanagi: I belong to the Laboratory of Genome Sciences in the Division of Bioengineering and Bioinformatics, where we study genomes from the standpoint of evolution in order to understand the relationship of genomes and phenotypes (disease, physical characteristics, etc.) and the diversity of living organisms. We are also developing different tools for use in genome research.
Studying genomes from an evolutionary standpoint involves comparing genomes of various living organisms and elucidating what kinds of effects their resemblances and differences have on their evolution. For example, we compare the sequences of particular genes between humans and chimpanzees, determine differences and when they have arisen, and infer how the differences affect the phenotypes of the two species.
Also, even within the same species, genes undergo subtle changes as they are passed on from generation to generation. Since some of these changes are believed to enhance an organism’s ability to withstand changing environments and to survive, comparing genes of past and present organisms would enable us to discover which genes play a role in enhancing their capacity to adapt to their environments. We are carrying out statistical analysis of genome information in genome databases (Ex. 1), but to compare genes that are not found in databases, we use the DNA sequencer (machine for automatic nucleotide sequencing) at our laboratory to sequence genes from a certain sample of an organism.
Discovery and classification of new types of virus causing “pink eye”
Can you give examples of research achievements made in your lab?
Dr. Koyanagi: An example of a recent topic is our collaborative research with the School of Medicine on adenoviruses. Adenoviruses cause pneumonia, conjunctivitis, and other infectious diseases, and include those types characterized by a high level of virulence, such as the one causing “pink eye” (epidemic keratoconjuctivitis), which is prevalent during the warm season. In our research, comparison of entire genome sequences of new types of adenovirus discovered in Japan with those of existing adenoviruses revealed that multiple genetic recombinations with other adenoviruses have occurred in the new species (Fig. 1). We are now conducting further research to determine whether the number and location of the recombinations are related to the virulence of the virus.
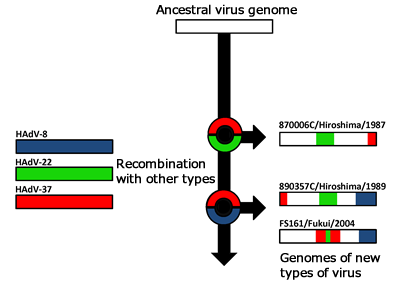
Evolution of genomes of new types of adenoviruses: Colored parts are regions in the genome believed to have undergone recombinations with other types. Circles denote recombination events.
Moving towards the next step in an environment that allows access to diverse areas of research
What kind of research themes do you plan to pursue in the future?
Dr. Koyanagi: With regard to adenoviruses, for example, we have been analyzing virus samples isolated in different periods from research institutions all over Japan to track the genetic changes that have taken place since the 1980s until the present time. We would also like to compare with samples from overseas and analyze geographical and other differences. Through this research, we hope to identify the particular region in the genome responsible for disease development and the high virulence of the Japanese strains. This is an example of how we would like to pursue research to understand the relationship of genomes and phenotypes from the standpoint of evolution.
Activities in the lab cover a wide variety of fields, including research on human and rice genomes and exploration of new deep-sea species (Ex. 2, Photo 1)). I think one of the attractive features of genome research is being able to study various organisms and life phenomena. Although genome research covers a wide range of topics, the Graduate School of Information Science and Technology gathers under one roof faculty coming from varied backgrounds, not only in engineering, but also in the sciences, agriculture, and other fields. We also have access to computer-based analysis methods, such as image processing and statistical analyses. Therefore, I think we can say that we have the best environment for conducting research involving large-scale data. Research that has not been possible ten years ago can now be possible with the current technology and knowledge at our disposal.

Explanation
Explanation 1. Genome database
Many databases on genome information analyzed by universities and research institutions around the world can be found on the Internet. Efforts are also being made at constructing integrated databases for the efficient use of these different databases. An example is the Integrated Database of Annotated Human Genes (H-InvDB), which is an integrated database of human genes and transcripts. Through this database, extensive analyses of all human transcripts are carried out to comprehend gene structure and other features of human genes and provide curated annotations of human genes and transcripts.
Explanation 2. New deep-sea species exploration project
Under a collaborative research project with the School of Fisheries, we take samples of mud from the deep seabed and analyze genetic information from various living organisms isolated from the mud to explore new species.

